Table of Links
3.1. Benefits scale with model size and 3.2. Faster inference
3.3. Learning global patterns with multi-byte prediction and 3.4. Searching for the optimal n
3.5. Training for multiple epochs and 3.6. Finetuning multi-token predictors
3.7. Multi-token prediction on natural language
4. Ablations on synthetic data and 4.1. Induction capability
5. Why does it work? Some speculation and 5.1. Lookahead reinforces choice points
5.2. Information-theoretic argument
7. Conclusion, Impact statement, Environmental impact, Acknowledgements, and References
A. Additional results on self-speculative decoding
E. Additional results on model scaling behavior
F. Details on CodeContests finetuning
G. Additional results on natural language benchmarks
H. Additional results on abstractive text summarization
I. Additional results on mathematical reasoning in natural language
J. Additional results on induction learning
K. Additional results on algorithmic reasoning
L. Additional intuitions on multi-token prediction
5. Why does it work? Some speculation
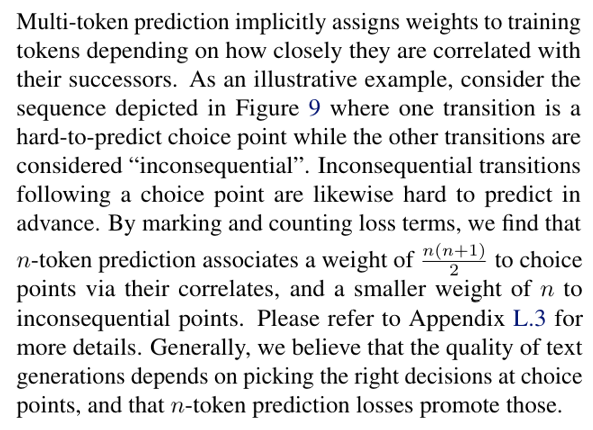
Why does multi-token prediction afford superior performance on coding evaluation benchmarks, and on small algorithmic reasoning tasks? Our intuition, developed in this section, is that multi-token prediction mitigates the distributional discrepancy between training-time teacher forcing and inference-time autoregressive generation. We support this view with an illustrative argument on the implicit weights multi-token prediction assigns to tokens depending on their relevance for the continuation of the text, as well as with an information-theoretic decomposition of multi-token prediction loss.
5.1. Lookahead reinforces choice points
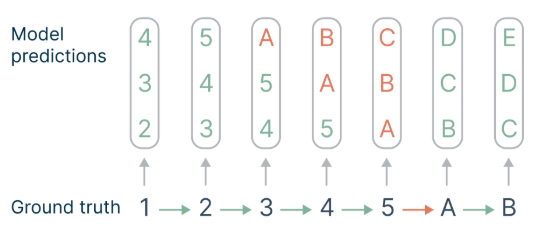
Not all token decisions are equally important for generating useful texts from language models (Bachmann and Nagarajan, 2024; Lin et al., 2024). While some tokens allow stylistic variations that do not constrain the remainder of the text, others represent choice points that are linked with higher-level semantic properties of the text and may decide whether an answer is perceived as useful or derailing.
This paper is available on arxiv under CC BY 4.0 DEED license.
Authors:
(1) Fabian Gloeckle, FAIR at Meta, CERMICS Ecole des Ponts ParisTech, and contributed equally;
(2) Badr Youbi IdrissiFAIR at Meta, LISN Université Paris-Saclay, and contributed equally;
(3) Baptiste Rozière, FAIR at Meta;
(4) David Lopez-Paz, FAIR at Meta and his the last author;
(5) Gabriel Synnaeve, FAIR at Meta and the last author.